We are interested in understanding how epigenetic marks are placed, read and interpreted on chromatin. Chromatin becomes decorated with post-translational modifications to control the myriad of DNA-related processes in the cell. We create modified chromatin using chemical biology and biochemical methods. We then use our defined modified chromatin to study individual nucleosome-chromatin protein complexes using single-particle cryo-electron microscopy (cryo-EM), Biochemical, Biophysical and Cell Biology approaches to investigate histone marks and DNA methylation.
How is DNA Methylation guided by chromatin?
DNA methylation is a common epigenetic mark that is often associated with turning off genes and compacting DNA. Other epigenetic marks have the power to regulate DNA methylation, controlling when and where DNA methylation is placed on DNA, but we do not understand how this works. We are rebuilding the DNA methylation machinery within chromatin to help us answer this question.
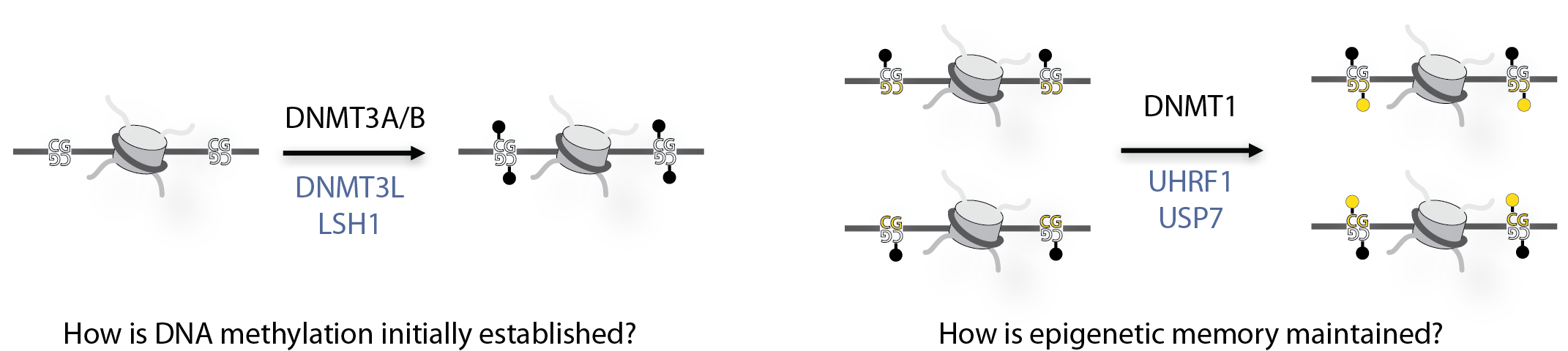
DNA methylation is a highly regulated process, so by looking at the structure of the methylation machinery and the modified nucleosomes we hope to understand how methylation is targeted at specific times and to specific sites on DNA, hopefully helping us to understand how this process can become faulty leading to disease.
How do post-translational modifications foster DNA repair?
DNA is under constant attack, which can cause unwanted genetic mutations and cancer. Luckily our cells have a host of DNA repair proteins, which help to fix most of the damage. These highly efficient repair proteins are recruited to sites of damage by recognition of DNA damage-specific marks on chromatin. We are hoping to understand how DNA damage is signaled on chromatin and how this leads to correct repair.
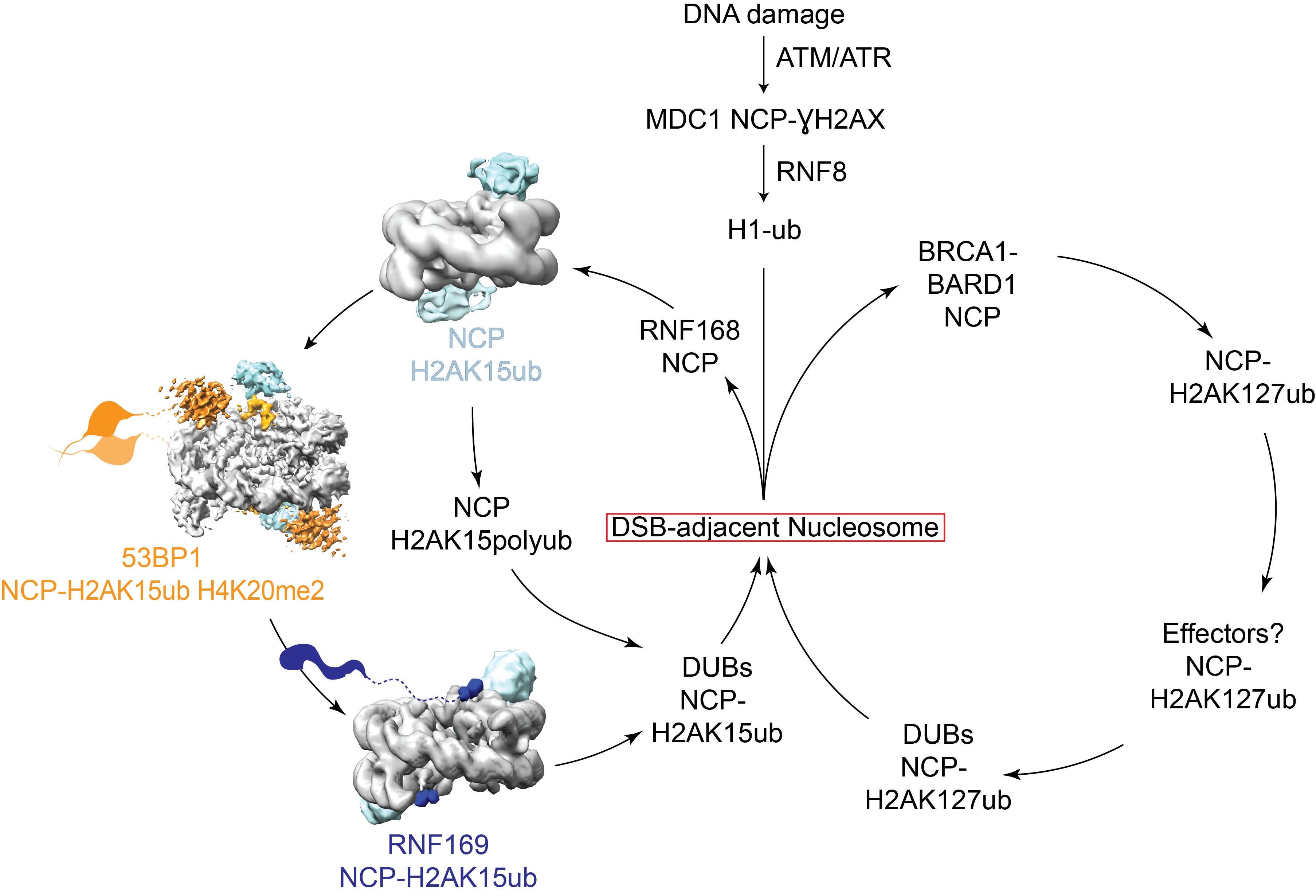
Nucleosome modifications act as a central signaling hub in this network to organise responses to a neighbouring break. While many factors are known to localise to modified DSB-adjacent chromatin, the exact function and binding of many of these proteins is unclear. We plan to focus on biochemically and structurally characterising nucleosome ubiquitylation proteins involved in DNA damage repair.
How is CHROMATIN ORGANISED IN DIVERSE EUKARYotes?
Publications
2026
- Trypanosome histone variants H3.V and H4.V promote nucleosome plasticity in repressed chromatin.
Deák G, Burdett H, Watson JA, Wilson MD.
Structure
bioRxiv Pubmed
2025
- Parasite nucleosomes: Chromatin dynamics rewired.
Deák G, Wilson MD.
PLoS Pathogens
Pubmed - Nucleosome interaction of the CPC secures centromeric chromatin integrity and chromosome segregation fidelity.
Gireesh A, Abad Fernandez M, Nozawa R, Sotelo-Parrilla P, Dury LC, Likhodeeva M, Spanos C, Cardenal Peralta C, Rappsilber J, Hopfner KP, Wilson MD, Vanderlinden WD, Hirota T, Jeyaprakash AA.
EMBO Journal
bioRxiv Pubmed - MeCP2 requires interactions with nucleosome linker DNA to read chromatin DNA methylation.
Watson JA, Alexander-Howden BK, Hall TS, Wear MA, McGhie F, Clifford G, Wapenaar H, Zou J, Adrian Bird A, Wilson MD.
bioRxiv - The kinetoplastid kinetochore protein KKT23 acetyltransferase is a structural homolog of GCN5 that acetylates the histone H2A C-terminal tail.
Ludzia P, Ishii M, Deák G, Spanos C, Wilson MD, Redfield C, Akiyoshi B.
Structure.
bioRxiv Pubmed
2024
- The N-terminal region of DNMT3A engages the nucleosome surface to aid chromatin recruitment.
Wapenaar H, Clifford G, Rolls W, Pasquier M, Burdett H, Zhang Y, Deák G, Zou J, Spanos C, Taylor MRD, Mills J, Watson JA, Kumar D, Clark R, Das A, Valsakumar D, Bramham J, Voigt P, Sproul D, Wilson MD.
EMBO Reports
bioRxiv Pubmed - Using human disease mutations to understand de novo DNA methyltransferase function.
Rolls W, Wilson MD, Sproul D.
Biochem Soc Trans.
Pubmed - DNMT3B PWWP mutations cause hypermethylation of heterochromatin.
Taglini F, Kafetzopoulos I*, Rolls W*, Musialik KI, Lee HY, Zhang Y, Marenda M, Kerr L, Finan H, Rubio-Ramon C, Gautier P, Wapenaar H, Kumar D, Davidson-Smith H, Wills J, Murphy LC, Wheeler A, Wilson MD¶, Sproul D¶.
EMBO Reports
bioRxiv Pubmed
2023
- BRCA1-BARD1 combines multiple chromatin recognition modules to bridge nascent nucleosomes.
Burdett H*, Foglizzo M ¶*, Musgrove LJ, Kumar D, Clifford G, Campbell LJ, Heath GR, Zeqiraj E¶, Wilson MD¶.
Nucleic Acids Research
bioRxiv Pubmed - Histone divergence in trypanosomes results in unique alterations to nucleosome structure.
Deák G, Wapenaar H, Sandoval G, Chen R, Taylor MRD, Burdett H, Watson JA, Tuijtel MW, Webb S, Wilson MD.
Nucleic Acids Research
bioRxiv Pubmed
2022
- Pore dynamics and asymmetric cargo loading in an encapsulin nanocompartment.
Ross J, McIver Z, Lambert T, Piergentili C, Gallagher KJ, Bird JE, Cruickshank FL, Zarazúa-Arvizu E, Horsfall LE, Waldron KJ, Wilson MD, Mackay LC, Baslé A, Clarke DJ, Marles-Wright J.
Science Advances
bioRxiv Pubmed
2021
- BARD1 reads H2A lysine 15 ubiquitination to direct homologous recombination.
Becker JR, Clifford G, Bonnet C, Groth A, Wilson MD, Chapman JR.
Nature
bioRxiv Pubmed
2020
- PALB2 chromatin recruitment restores homologous recombination in BRCA1-deficient cells depleted of 53BP1.
Belotserkovskaya R¶, Raga Gil E, Lawrence N, Butler R, Clifford G, Wilson MD¶, Jackson, SP¶ .
Nature communications
Pubmed - The Chaperone FACT and Histone H2B Ubiquitination Maintain S. pombe Genome Architecture through Genic and Subtelomeric Functions.
Murawska M, Schauer T, Matsuda A, Wilson MD, Pysik T, Wojcik F, Muir TW, Hiraoka Y, Straub T, Ladurner AG.
Molecular Cell
Pubmed
2019
- MDC1 PST-repeat region promotes histone H2AX-independent chromatin association and DNA damage tolerance.
Salguero I, Coates J, Belotserkovskaya R, Sczaniecka-Clift M, Demir M, Jhujh S, Wilson MD, Jackson SP.
Nature Communications
Pubmed - Retroviral integration into nucleosomes through DNA looping and sliding along the histone octamer.
Wilson MD*, Renault L*, Maskell D, Ghoneim M, Pye VE, Nans N, Rueda DS, Cherepanov P, Costa A.
Nature Communications
bioRxiv Pubmed - Analysis of RNA polymerase II ubiquitylation and proteasomal degradation.
Vidakovic AT, Harreman M, Dirac-Svejstrup AB, Boeing S, Roy A, Encheva V, Neumann M, Wilson MD, Snijders AP, Svejstrup JQ.
Methods
Pubmed
2018
- Inhibition of 53BP1 favors homology-dependent DNA repair and increases CRISPR-Cas9 genome-editing efficiency.
Canny MD, Moatti N, Wan LCK, Fradet-Turcotte A, Krasner D, Mateos-Gomez PA, Zimmermann M, Orthwein A, Juang YC, Zhang W, Noordermeer SM, Seclen E, Wilson MD, Vorobyov A, Munro M, Ernst A, Ng TF, Cho T, Cannon PM, Sidhu SS, Sicheri F, Durocher D.
Nat Biotechnology
PubMed
2017
- Reading chromatin signatures after DNA double-strand breaks.
Wilson, MD¶, and Durocher, D.
Philos Trans R Soc Lond B Biol Sci .
PubMed - The RNF168 paralog RNF169 defines a new class of ubiquitylated histone reader involved in the response to DNA damage.
Kitevski-LeBlanc J, Fradet-Turcotte A, Kukic P*, Wilson MD*, Portella G, Yuwen T, Panier S, Duan S, Canny MD, van Ingen H, Arrowsmith CH, Rubinstein JL, Vendruscolo M, Durocher D, Kay LE.
Elife
PubMed - Cryo-electron microscopy of chromatin biology.
Wilson, MD¶, and Costa, A¶.
Acta Crystallogr D Struct Biol.
PubMed
2016
- The structural basis of modified nucleosome recognition by 53BP1.
Wilson, MD*, Benlekbir, S*, Fradet-Turcotte, A, Sherker, A, Julien, JP, McEwan, A, Noordermeer, SM, Sicheri, F, Rubinstein, JL, and Durocher, D.
Nature
PubMed
2013
- Proteasome-mediated processing of Def1, a critical step in the cellular response to transcription stress.
Wilson MD*, Harreman M*, Taschner M, Reid J, Walker J, Erdjument-Bromage H, Tempst P, and Svejstrup JQ. (2013).
Cell
PubMed - Ubiquitylation and degradation of elongating RNA polymerase II: the last resort.
Wilson MD, Harreman M, and Svejstrup JQ.
Biochim Biophys Acta.
PubMed
2012
- MultiDsk: a ubiquitin-specific affinity resin.
Wilson MD, Saponaro, M, Leidl MA, and Svejstrup JQ.
PLoS One
PubMed
2011
- The PI3K p110delta regulates expression of CD38 on regulatory T cells.
Patton DT, Wilson MD, Rowan WC., Soond DR, and Okkenhaug K.
PLoS One
PubMed
* authors contributed equally ¶ co-corresponding author(s)